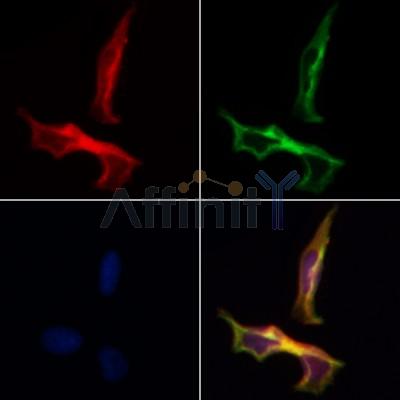
产品描述
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: 适用于变性蛋白样本的免疫印迹检测. IHC: 适用于组织样本的石蜡(IHC-p)或冰冻(IHC-f)切片样本的免疫组化/荧光检测. IF/ICC: 适用于细胞样本的荧光检测. ELISA(peptide): 适用于抗原肽的ELISA检测.
引用格式: Affinity Biosciences Cat# AF0097, RRID:AB_2833278.
展开/折叠
14 3 3 delta; 14 3 3 protein zeta/delta; 14 3 3 protein/cytosolic phospholipase A2; 14 3 3 zeta; 14-3-3 protein zeta/delta; 1433Z_HUMAN; Epididymis luminal protein 4; Epididymis secretory protein Li 3; HEL S 3; HEL4; KCIP-1; KCIP1; MGC111427; MGC126532; MGC138156; Phospholipase A2; Protein kinase C inhibitor protein 1; Tyrosine 3 monooxygenase/tryptophan 5 monooxygenase activation protein, delta polypeptide; Tyrosine 3 monooxygenase/tryptophan 5 monooxygenase activation protein, zeta; Tyrosine 3 monooxygenase/tryptophan 5 monooxygenase activation protein, zeta polypeptide; Tyrosine 3/tryptophan 5 monooxygenase activation protein, zeta polypeptide; YWHAD; YWHAZ; 14 3 3 alpha; 14 3 3 protein beta/alpha; 14-3-3 protein beta/alpha; 1433B_HUMAN; Brain protein 14 3 3 beta isoform; GW128; HS 1; KCIP-1; KCIP1; N-terminally processed; Protein 1054; Protein kinase C inhibitor protein 1; YWHAA; YWHAB;
抗原和靶标
A synthesized peptide derived from human 14-3-3 zeta/delta, corresponding to a region within the internal amino acids.
研究领域
· Cellular Processes > Cell growth and death > Cell cycle. (View pathway)
· Cellular Processes > Cell growth and death > Oocyte meiosis. (View pathway)
· Environmental Information Processing > Signal transduction > PI3K-Akt signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Hippo signaling pathway. (View pathway)
· Human Diseases > Infectious diseases: Bacterial > Pathogenic Escherichia coli infection.
· Human Diseases > Infectious diseases: Viral > Hepatitis B.
· Human Diseases > Infectious diseases: Viral > Epstein-Barr virus infection.
· Human Diseases > Cancers: Overview > Viral carcinogenesis.
限制条款
产品的规格、报价、验证数据请以官网为准,官网链接:www.affbiotech.com | www.affbiotech.cn(简体中文)| www.affbiotech.jp(日本語)产品的数据信息为Affinity所有,未经授权不得收集Affinity官网数据或资料用于商业用途,对抄袭产品数据的行为我们将保留诉诸法律的权利。
产品相关数据会因产品批次、产品检测情况随时调整,如您已订购该产品,请以订购时随货说明书为准,否则请以官网内容为准,官网内容有改动时恕不另行通知。
Affinity保证所销售产品均经过严格质量检测。如您购买的商品在规定时间内出现问题需要售后时,请您在Affinity官方渠道提交售后申请。产品仅供科学研究使用。不用于诊断和治疗。
产品未经授权不得转售。
Affinity Biosciences将不会对在使用我们的产品时可能发生的专利侵权或其他侵权行为负责。Affinity Biosciences, Affinity Biosciences标志和所有其他商标所有权归Affinity Biosciences LTD.